
::home:: ::progress:: ::partners:: ::training:: ::about in3:: .:ESTIV home:.
In3 Project update – Jan 2021
Update on the in3 project: An integrated interdisciplinary approach to animal-free nanomaterial and chemical safety assessment – the Corona year…
Dear ESTIV members! It is with great pleasure, that we update you on the in3 activities that have taken place in the previous, memorable year 2020. As a short reminder, the MSCA-ITN in3 employs 15 PhD candidates, most of which by now have finished the 36 months of intensive training and research. These highly motivated and dedicated scientists from all over the world are hosted in laboratories across Europe, from the UK to Hungary. Their ultimate goal is the progression of non-animal methods and the integration of in silico and in vitro tools for toxicity assessment. ESTIV is one of our partner organisations and hosts our web page (https://estiv.org/in3/).
With the in3 project almost at the end, the in3 ESRs have turned into extensively trained in vitro and in silico toxicologists. The focus in this past year was on the research output, finalization of secondments, (transcriptomics) data analysis and manuscripts writing. These activities will be wrapped up in the next six months as the project was granted an extension till the end of June 2021.
Slovenia and Amsterdam meetings
In February 2020 Marjana Novič from the Kemijski Inštitut in the beautiful, eco-friendly and hospitable city of Ljubljana, Slovenia, hosted the in3 project workshop and the 5th Network meeting. Luckily this was just organized before labs were closed and international traveling was stopped due to COVID-19 regulations. Maybe the winter storms that prevented the Amsterdam group to fly out on the Sunday afternoon were a token that a different and difficult year was coming…?

From left to right front row; Ana Caballero, Emilio Benfenati, Nicola Minovski, Andras Dinnyes, Walter Pfaller, Vidya Chandrasekaran, Pranika Singh, Carolina Nunes, Zahra Mazidi, Susana Proenca, Liadys Morales, Leonie Fransen, Anja Wilmes, Nicoleta Spinu. From left to right top row: Ellen Langemeijer, Mark Cronin, Giovanni Grillari, Thomas Exner, Kristijan Vukovic, Cormac Murphy, Sara Wellens, Maxime Culot, Paul Jennings, Sreya Ghosh, Ivo Djidrovski, Marjana Novic (KI host), Nynke Kramer, Melinda Zana.
Anyway…. The Workshop on data analysis and management provided by the bioinformaticians from in3 partner Edelweiss Connect, Thomas Exner and Pranika Singh, familiarized the in3 ESRs with the transcriptomics data analysis workflow. Approximately 3000 transcriptomics samples were run from several different iPSC-derived in vitro systems exposed to different test compounds at non cytotoxic concentrations. This joint analysis will provide us with knowledge on the Mode of toxic Action of these compounds.

Workshop attendees working with the Edelweiss connect provided TempO-Seq data analysis Workflow.
During the spring and summer of 2020, the transcriptomics data meta-tagging was completed, data was normalised, and the matured in3 transcriptomics workflow was established. Control samples (0.1 % DMSO treated) were compared, to undifferentiated iPSC cells and compound exposed differentiated cells like hepatocytes, neuronal spheres, endothelial cells, podocytes and proximal tubular like (renal) cells. Pathway annotation lists for toxicity related pathways were created by Vidya and Sara, and the treatment- and cell comparisons took off. With the fantastic online help from colleagues like Thomas Exner and Pranika Singh, the in3 data analysis continued during lock-down.
Secondments planned in 2020 for the ESRs that didn’t fulfil these earlier, were hindered by the COVID-19 lockdown. In March, two ESRs had to relocate to their home countries due to lab closures. Several planned secondments were cancelled and online secondments were started as a suitable alternative. With all the data analysis that was needed at that time anyway, the timing of the lock down for in3 wasn’t the worst, but took some time for the ESRs to adjust their schedules.
A TIV special issue was launched in the fall of 2020 in order to offer a one-stop-shop for the in3 transcriptomics analysed data. Manuscripts are currently drafted and will be finalised in the coming months/year. The in3 partner NewCells Biotech shifted focus slightly upon receiving the confirmation that their iPSCs derived lung cells express the ACE-2 receptor, which is highly relevant in coronavirus uptake in cells. COVID-19 research might benefit from the developed iPSCs derived lung model developed by the in3 ESR at NCB.
Training of the in3 ESRs ended in April 2020 with the online Ethics in Science course, organized by Maxime Culot, which was scored Excellent by the ESRs. Divers aspects on ethical behaviour from scientists were discussed in the common, informal in3 setting. Testing of background knowledge via anonymous polls indicated that not every ethical questionable situation is as simple and clear as one might think, and that communication and discussion is always required to reach a consensus on steps to undertake…
Mid-December 2020 the in3 workshop on Career Development and the 6th network meeting were organized virtually via the in3 (estiv.org) website. To facilitate communication during the online meeting a dedicated Slack (chat) workspace was created.
On Monday 14 December, a line-up of excellent toxicologists from regulatory body (Sandra Coecke, EURL ECVAM), publishing house (Georgina Harris, Frontiers) and various industries (Manon Beekhuijzen, CRL; Carl Westmoreland, Unilever; Ashwani Sharma, Senzagen and Samuel Constant, Epithelix) presented their career path and discussed various aspects on careers with the ESRs. The offered advice and suggested routes to go for a brilliant career were welcomed enthusiastically.
A relaxing evening followed. The live music session from the European all female band “Panienki” featuring Thomas helped very much to unwind, and dancing made us all longing for meeting in person again!
The presentation of scientific progress on Tuesday 15 December was done via pre-recorded lectures that were posted online via the in3 website. Each ESR was introduced by his/her supervising PI in a movie. In combination with the questions posted in the Slack chat, the dedicated discussion slots with the ESRs provided ample opportunity to dive into the scientific progress made. During the evening, an online pub quiz topped off with dancing to online music requests closed of this never-a-dull-moment 6th in3 meeting.
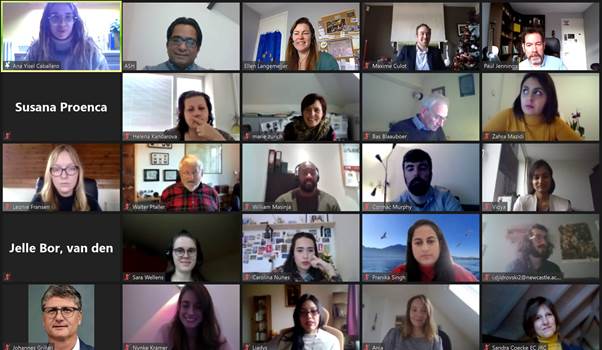
Part of the attendees during the online in3 meeting in Career Development on14 December 2020.
We also had some interesting scientific publications this year and expect many more to come. You will be informed when the TIV special edition is available.
- Quantitative adverse outcome pathway (qAOP) models for toxicity prediction. Spinu, N., Cronin, M., Enoch, S. J., Madden, J. C., & Worth, A. P. (2020). Archives of toxicology, 94(5), 1497–1510. https://doi.org/10.1007/s00204-020-02774-7
- Homology Modeling of the Human P-glycoprotein (ABCB1) and Insights into Ligand Binding through Molecular Docking Studies. Mora Lagares L, Minovski N, Caballero Alfonso AY, Benfenati E, Wellens S, Culot M, Gosselet F, Novič M. Int J Mol Sci. 2020 Jun 5;21(11):4058. doi: 10.3390/ijms21114058. PMID: 32517082
- See the in3 website for the overview of in3 publications; in3 (estiv.org)
in3 Disseminations in 2020 were limited due to the COVID-19 related lockdown. In fact, the EUSAAT conference in Linz, Austria (10th to 13th of October, 2019) was the final conference in which an in3 delegation participated. Unfortunately, the SOT2020, QSAR2020 and ESTIV meeting in Sitges, Spain June 2020, were cancelled or postponed.

Paul, Marie-Gabrielle, Maxime (PIs), and Zahra, Sara and Carolina during the EUSAAT conference, Linz, October 2019.

ESR Ivo Djidrovski, working on an iPSC derived lung model presented his research in October 2019 at the Association of Inhalation Toxicologists, Brighton, UK as well as in the House of Commons (London, UK) early March 2020.
As off fall 2020 online dissemination of the open source in silico tools developed by in3 are available via the VEGA websites. VEGA accommodates thousands of users (scientists who download the software) and is linked to the OECD QSAR Toolbox, ECHA to prioritize substances, as one of the three examples of software in their document “Practical guide on How to use and report QSAR”, and mentioned in the EFSA Guidance documents on Weight of Evidence and Chemical Mixtures. Additionally, VEGA is used within the AMBIT platform developed by CEFIC for Read across of Chemical substances.
A final in3 dissemination meeting will be organized in June 2021, most likely online. Alternatively, you can find us at the in3 session (online or in person, fingers crossed 😉) during the upcoming 11th World Congress on Alternatives and Animal Use in the Life Sciences (WC11) rescheduled to 22-26 August 2021, Maastricht, NL.
Thank you for your interest. Keep an eye on our webpage http://estiv.org/in3/
or follow us on twitter @in3itn.
All the best,
Dr. Ellen Langemeijer, in3 project manager
Prof. Dr. Paul Jennings, in3 coordinator
Project 1: Utilization of human iPSC derived renal proximal tubular epithelial-like cells for understanding the role of megalin transporter in nephrotoxicity
|
ESR: Vidya
Chandrasekaran Nationality: Indian Host Organization: Vrije Universiteit, Amsterdam Person in charge: Prof. Dr. Paul Jennings email: p.jennings@vu.nl; v.chandrasekaran@vu.nl |
 |
Vidya is involved in developing proximal tubular-like epithelial cells (PTL) from induced Pluripotent Stem Cells (iPSC) obtained from different donors. The expression of PT specific marker, megalin (brush border protein), transporters and the epithelial architectures were tested to be positive in these developed PTL cells. Furthermore, it was subjected to test for the functionality of the megalin system by examining their sensitivity to gentamicin. As the next step, further characterization of the PTL for the functionality of transporters and expression of PT markers will be done. Also, the role of megalin/cubilin system in chemical induced nephrotoxicity will be investigated deeply using the PTL cells by incorporating toxicodynamic and toxicokinetic studies. This will as well help us develop Adverse Outcome Pathway.
Potential outcomes:
- An optimised iPSC derived PT differentiation protocol characterised with respect to xenobiotic handling and tissue specific expression and to challenge the models with nephrotoxins
- Investigation of donor specific effects
- Investigation of megalin/cubilin system mediated nephrotoxicity of chemicals and nanoparticles.
- To challenge the optimised system to the in3 compounds and perform mechanistic and kinetic studies
- Deployment of reporter iPSC and testing it with a larger compound set
- To create and challenge a renal AOP with PT cells as the initiating / affected cell
Project 2: Generation of a glomerular model from iPSC and application to nephrotoxicity testing
|
ESR: Cormac Murphy Nationality: Irish Host Organization: Vrije Universiteit, Amsterdam Person in charge: Prof. Paul Jennings email: p.jennings@vu.nl; c.murphy@vu.nl |
 |
Cormac is involved in differentiating induced pluripotent stem cells (iPSC) using a modified, one-step differentiation protocol, developed from the one used by Song et al. 2012. Cormac has generated podocyte-like cells that exhibit the characteristic podocyte foot processess. It was also checked for the expression of podocyte-associated markers podocin, synaptopodin, nephrin and WT1 using immunocytochemistry. The next step will be to co-culture the podocytes with endothelial cells, derived from iPSCs from the same donors, and encourage the formation of a glomerular basement membrane between them, thus completing the three main components of the glomerular filtration barrier. This entirely iPSC derived glomerular model will then be challenged with the in3 test compounds to gain more information on the mechanisms of glomerular toxic reactions and aid in the development of Adverse Outcome Pathways for future regulator assessment.
Potential outcomes:
- A glomerular model from iPSC incorporating podocyte-like cells and autologous endothelial cells which can be used to investigate compound / nanomaterial glomerular filtration rates and also predict glomerular toxins
- Investigation of donor specific effects utilising iPSC from donors with monogenetic glomerular disorders
- To expose the glomerular models to in3 test compounds for mechanistic and kinetic analysis and modeling
- To challenge the optimised system to the in3 compounds and perform mechanistic and kinetic studies
- To create and challenge a renal AOP with the glomerulus as the initiating or affected site
Project 3: iPSC derived endothelial cells
|
ESR: Zahra Mazidi Nationality: Iranian Host Organization: Evercyte GmbH, Vienna, Austria Person in charge: Prof. Regina Grillari email: Regina.Grillari@evercyte.com; Zahra.Mazidi@evercyte.com |
 |
Zahra is involved in differentiating endothelial cells (ECs) from induced Pluripotent Stem Cells (iPSC) obtained from different donors (one male and one female). She succeeded in establishing protocols to differentiate iPSCs versus ECs. Cells can be banked for further use at later stages at two steps within this differentiation protocol, as endothelial colony forming cells (Vegfr2+), and mature endothelial cells (CD31+). Currently, she is characterizing the functionality of these iPSC derived endothelial cells. The exact mechanism of toxicity on the cells is studied using the 10 selected toxic chemicals, starting with cyclosporin A. As the next step, she will develop endothelial reporter cell lines targeting toxic relevant promoter genes. All this data together will support the development of at least one Adverse Outcome Pathway (AOP) for endothelial cells. Eventually, using these tools, she will have a suitable in vitro model for vascular toxicity studies that due to the source of iPSCs that can be generated from any individual, can be used as a path towards personalized toxicity studies.
Potential outcomes:
- A generic donor-matched endothelial model with specific characteristics and responses
- Investigation of donor specific effects utilising iPSC from donors
- To expose the endothelial model to in3 test compounds for mechanistic and kinetic analysis and modelling
- To create and challenge a vascular AOP
- To create endothelial co-cultures with kidney, liver and lung systems and characterised altered endothelial phenotypes due to cross talk
Project 4: Differentiation of brain like endothelial cells from iPSC and application to BBB toxicity testing and prediction of CNS distribution
|
ESR: Sara Wellens Nationality: Belgian Host Organization: University of Artois, Lens, France Person in charge: Ass. Prof. Maxime Culot email: maxime.culot@univ-artois.fr; sara_wellens@ens.univ-artois.fr |
 |
Sara is involved in establishing in vitro BBB models from the iPSCs of interest by evaluating different published protocols to differentiate iPSCs into BBB like endothelial cells regarding their expression of endothelial markers, formation of tight barrier and the presence of functional efflux pumps.. This project will involve the optimisation, characterisation and utilisation of induced pluripotent stem cells for the derivation of donor specific podocyte cells. Work will involve cell culture, molecular biology approaches, in vitro toxicity assays and mechanistic toxicology including transcriptomics.
Potential outcomes:
- An iPSC derived BBB differentiation protocol suitable for both toxicity testing and transport studies characterised with respect to xenobiotic handling and tissue specific expression will be challenged with toxic compounds to identify the window of opportunity for exposure
- Investigation of donor specific effects
- To challenge the optimised system to the in3 compounds and perform mechanistic and kinetic studies
- Deployment of reporter iPSC and testing it with a larger compound set
- In collaboration with ERS5 to create a brain AOP and use the mechanistic and kinetic data to challenge and refine the AOP
Project 5: Differentiation of brain cells from iPSC and application to neurotoxicity testing
|
ESR: Carolina Nunes Nationality: Portuguese Host Organization: University of Lausanne, Lausanne, Switzerland Person in charge: Dr. Marie Gabrielle Zurich email: Marie-Gabrielle.ZurichFontanellaz@unil.ch; carolina.nunes@unil.ch |
 |
Carolina is establishing the 3D human brain model that was proposed by Pamies et al, 2017. Briefly, the differentiation of iPSCs into neuroprogenitor cells (NPCs) is induced in 2D culture by using a commercially available medium. Then, the differentiation of NPCs into brain cells is driven by another culture medium, containing trophic factors. Characterization of NPCs and 3D cultures is performed by qRT-PCR, immunohistochemistry and FACS analyses. Results shows the quality of NPCs, as well as the presence of neurons, astrocytes and oligodendrocytes in the 3D cultures. Carolina is involved in developing a protocol to add functional and stable iPSC-derived microglia to the current 3D system in order to obtain a model able to produce a neuroinflammatory response upon toxic exposure. This model will then be used to evaluate neurotoxicity of the in3 compounds, and to develop new quantitative Adverse Outcome Paths.
Potential outcomes:
- An iPSC derived 3D model of the brain suitable for neurotoxicity testing including evaluation of inflammatory response upon toxicant exposure
- Investigation of donor specific effects
- To challenge the optimised system to the in3 compounds and perform mechanistic and kinetic studies
- Deployment of reporter iPSC and testing it with a larger compound set
- In collaboration with Sara (ERS4) to create a brain AOP and use the mechanistic and kinetic data to challenge and refine the AOP
Project 6: Generation of Fluorescent reporter induced pluripotent stem cells (iPSCs)
|
ESR: Aurore Bourguignon Nationality: French Host Organization: Biotalentum, Gödöllö, Hungary Person in charge: Prof. Andras Dinnyes email: andras.dinnyes@biotalentum.hu; aurore.bourguignon@biotalentum.hu |
 |
Aurore is involved in the incorporation of fluorescent constructs into selected iPSC lines, using CRISPR/Cas9 technology. This will generate fluorescent reporters of toxicity, which can be then be differentiated into toxicologically relevant tissues. A gene within the Nrf2 pathway has been initially targeted. Depending on the progress, multiple labels may be attempted, and other pathways investigated. Then the response of undifferentiated and differentiated iPSC to in3 compounds will be tested in order to compare the responses of the reporter and its parent lineage. The work will therefore involve biomolecular approaches, cell culture and the use of viability assays, stress assays, transcriptomics and kinetic assays.
Potential outcomes:
- Development of iPSCs reporter lines for at least one toxicity pathway (e.g. Nrf2 oxidative stress-induced pathway)
- To challenge the reporter iPSCs with the in3 compounds and compare these results to the parental iPSC lines
- Investigation of donor specific effects
- To transfer the reporters to the other laboratories and to monitor differentiation progress, then perform toxicity studies on these differentiated cells
- To carry out differentiation protocols in house and challenge these cells with a larger compound set in order to investigate transferability, reproducibility, and robustness of the protocols
Project 7: Differentiation of hepatocyte-like cells from induced pluripotent stem cells (iPSCs) and application to hepatotoxicity testing
|
ESR: Sreya Ghosh Nationality:Indian Host Organization: University of Leuven, Leuven, Belgium Person in charge: Prof. Catherine Verfaillie email: catherine.verfaillie@med.kuleuven.be; sreyahillol.ghosh@kuleuven.be |
 |
Sreya is involved in the improvement of protocols to derive hepatocyte-like cells (HLCs) from iPSCs using combinations of genome engineering to enhance/suppress key transcriptional networks, and achieve more advanced 2D and 3D cultures. These cultures will then be used to investigate uptake, extrusion and metabolism of model compounds and exposed to the in3 test compounds with cell viability measured. The model will also be used to develop the Adverse Outcome Pathway of NASH. Work will involve cell culture, molecular biology approaches, in vitro toxicity assays and mechanistic toxicology.
Potential outcomes:
- An iPSC derived hepatocyte differentiation protocol suitable for toxicity testing
- Investigation of donor specific effects, initially using donors with P-gp mutations
- Deployment of reporter iPSC and testing it with a larger compound set
- To create and challenge liver Adverse Outcome Pathways including steatosis
Project 8: Development of induced pluripotent stem cells (iPSCs)-derived small airway models and application to toxicity testing
|
ESR: Leonie Fransen Nationality: Dutch Host Organization: Public Health England, Centre for Radiation, Chemical and Environmental Hazards, Oxfordshire, United Kingdom Person in charge: Dr. Martin Leonard email: martin.leonard@phe.gov.uk; leonie.fransen@phe.gov.uk |
 |
Leonie is involved in the optimisation of iPSC-derived small airway models using published protocols. Small airway epithelium (SAE) will be cultured in a monolayer on TransWell inserts to allow for an air-liquid interface (ALI) culture. Macrophages are known to play an important role in small airway toxicity and the possibility to develop co-cultures with SAE will be investigated. Once the protocol is optimized, it will be used for toxicity testing of nanomaterials and in3 test compounds. The cells will be exposed using the VitroCell® cloud 6 system, allowing exposure to nebulized particles and chemicals. Toxicological effects on several differentiated iPSC cell lines will be compared to undifferentiated iPSC cells and primary cell lines to allow for the detection of cell type specific toxicity. This will be carried out using standard biochemical toxicity assays and transcriptional assessment using TempOseq technology. Ultimately these results will be integrated with AOP and in silico activities.
Potential outcomes:
- iPSC derived small airway differentiation protocols in an air-liquid interface suitable for toxicity testing
- Investigation of donor specific effects
- To challenge the optimised system to the in3 compounds and perform mechanistic and kinetic studies
- Deployment of reporter iPSC and testing it with a larger compound set
- To create a pulmonary Adverse Outcome Pathway by conducting a systematic review to identify hazards from chemical and nanomaterial inhalation exposure
Project 9: Development of iPSC derived Conducting Airways and application to toxicity testing
|
ESR: Ivo Djidrovski Nationality: Macedonian Host Organization: Newcells Biotech, Newcastle, United Kingdom Person in charge: Prof. Lyle Armstrong email: lyle.armstrong@newcastle.ac.uk; ivo.djidrovski@newcellsbiotech.co.uk |
 |
Ivo is involved in the optimisation of protocols for the derivation of donor specific conducting airway epithelium with the necessary multicellular components of ciliated epithelial cells, Clara cells, goblet cells and basal cells. To goal is to assess drug induced systemic lung toxicity. Two different protocols were tested (monolayer and spheroids) with two different iPSC lines (SBAD2 and SBAD3). The cells were differentiated during 45 days and characterized by immunostaining for specific markers. Both protocols gave satisfactory results with likely differentiation into ciliated cells, club cells, clara cells, goblet cells and alveolar type I and type II cells. As the next step, Improvement of differentiation protocol by sorting specific progenitor cells will be performed and optimised for the toxicity testing. Furthermore, the differentiated cells will be tested for the functionality of the cells. Once it is optimised, the cells will be challenged with chemical aerosols and nanoparticles in the air compartment.
Potential outcomes:
- An iPSC derived conducting airway differentiation protocol for systemic lung toxicity testing
- The model will be characterised with respect to drug and nanoparticle handling and tissue specific expression of ciliated epithelia, Clara, goblet, and basal cells. The model will be challenged with toxins and nanoparticles to investigate application to toxicity scenarios and to identify the window of opportunity for exposure
- Investigation of donor specific effects, initially using donors with P-gp SNPs
- Deployment of reporter iPSC and testing it with a larger compound set
- To create and optimise a pulmonary AOP in collaboration with ESR8 working on conducting airways this will involve a systematic review to identify hazards from chemical and nanomaterial inhalation exposure
Project 10: New in silico models for safer chemicals
|
ESR: Kristijan Vukovic Nationality: Croatian Host Organization: Istituto di Ricerche Farmacologiche Mario Negri, Milan, Italy Person in charge: Prof. Emilio Benfenati email: emilio.benfenati@marionegri.it; kristijan.vukovic@marionegri.it |
 |
Kristijan aims to develop improved in silico models for cosmetics and other substances, with reference to the endpoints of the project. The main goal is to establish a sound basis for the use of QSAR models for the assessment of chemicals, integrating the results several QSAR models for the same endpoint, and taking advantage, when useful, of the results of related endpoints, such as metabolism. Kristijan will acquire the practical skills for using the several QSAR models within different platforms, such as VEGA, EPISuite, Toxtree, TEST, OECD toolbox, CORAL, R and will also evaluate possible integration of different programs within a single platform. New models will be developed, applying software such as SARpy and QSARpy. Kristijan will assess the applicability domain of the best models for the chemicals which will be studied within in3 with the in vitro and in silico models. The results of the in vitro models addressed within in3 will be investigated, in order to extract rules for toxicity and implement them within VEGA (www.vega-qsar.eu).
Potential outcomes:
- New QSAR models with reference to AOPs will be developed, and implemented within VEGA
- Evaluation of other tools for use within the consortium for in vitro and in silico data integration
- Implementation of the tools for chemical safety assessment of project related compounds and models
Project 11: New read across modules for safer chemicals
|
ESR: Ana Yisel Caballero Nationality: Cuban Host Organization: Istituto di Ricerche Farmacologiche Mario Negri, Milan, Italy Person in charge: Prof. Emilio Benfenati email: emilio.benfenati@marionegri.it; ana.caballero@marionegri.it |
 |
The project aims to develop improved read-across modules, with reference to the endpoints of the project.To learn VEGA, SARpy, QSARpy, IstChemFeat, AMBIT, OECD Toolbox. The main goal is to establish a sound basis for the use of read-across for the assessment of chemicals, and then to integrate the results of the QSAR models with results from read-across. Ana will acquire the practical skills for using read-across, including the software such as ToxRead, AMBIT and OECD toolbox. New modules for read-across will be implemented within ToxRead (www.toxread.eu), and made freely available. The new innovative approach, suitable within a weight-of-evidence perspective, will be made available within the platform ToxRead (www.toxgate.eu).
Potential outcomes:
- New read-across modules which address the AOPs will be developed
- Evaluation of other tools for use within the consortium for in vitro and in silico data integration.
- Implementation of the tools for chemical safety assessment of project related compounds and models.
Project 12: Modelling of quantitative Adverse Outcome Pathways
|
ESR: Nicoleta Spînu Nationality: Romanian Host Organization: Liverpool John Moores University, Liverpool, United Kingdom Person in charge: Prof. Mark Cronin email: M.T.Cronin@ljmu.ac.uk; N.Spinu@2017.ljmu.ac.uk |
 |
Nicoleta is responsible for the modelling of the in3 Project's AOPs into quantitative AOPs (qAOPs). Pre-existing AOPs were identified by reviewing the Organisation for Economic Co-operation and Development (OECD) AOP Development Programme, specifically the AOP-Wiki Knowledge Base module. This exercise helped to formulate and map AOP networks that represent the complexity of biological and toxicological processes and therefore, the real-world exposure scenarios. The resultant AOP networks were analysed analytically and are used for further computational modelling. Future work covers the formulation of a general framework for qAOP development. Project data (kinetic and mechanistic) will be used to verify and optimise these AOPs and create quantitative relationships. The formalised qAOPs for different organ level toxicities will be placed in a mathematical model framework and coded with e.g. KNIME, R etc. Ultimately a software tool will be developed and combined with the read-across and QSAR models enabling input of both chemical structure and data from the assays for the key events. This integrated modelling approach will enable predictions to be made supported by both relevant chemistry and mechanistic biology.
Potential outcomes:
- Scoping of AOPs for kidney, liver, lung, brain and vasculature system
- Population of AOP constructs with data from existing sources
- Creation of qAOPs with project data
- Modelling mathematically the qAOPs with the possibility to predict potency
- Development of a software tool to combine kinetics, read-across and qAOPs
Project 13: P-glycoprotein modelling: New methodology of combining molecular modelling with in silico models for safer chemicals
|
ESR: Liadys Mora Lagares Nationality: Colombian Host Organization: National Institute of Chemistry, Ljubljana, Slovenia Person in charge: Prof. Marjana Novič email: Marjana.Novic@ki.si; liadys.moralagares@ki.si |
 |
Liadys is responsible of developing a methodology for improved in silico models including characterization of substrate specificity and P-gp transport of exogenous chemicals. The main goal is to develop in silico approaches that provide rapid and cost-effective screening platforms for the identification of P-gp substrates as well as for their transport characteristics. This will involve pharmacophore modelling and molecular docking aiming to predict P-gp inhibition properties of small drug molecules or pharmacologically-relevant P-gp substrates. This sub-project will combine the outcome of the molecular modelling results with the predictions resulting from QSAR models developed within the project. The in vitro studies performed by project partners will be utilized and compared with the in silico results to generate a two way optimisation.
Potential outcomes:
- Development of an in silico approach that provide a rapid and cost-effective screening platform for the identification and prediction of P-gp substrates, inhibitors and non-active compounds
- Identification of molecular interactions from molecular dynamics and docking studies of P-gp, with substrates relevant in cosmetics and other substances of concern within the project, in order to understand their possible transporting profile on molecular level
- Combination of QSAR prediction results with the molecular modelling outcome in order to integrate in vitro and in silico data
- Implementation of the tools for chemical safety assessment of project related compounds and models
Project 14: In vitro and in vivo biokinetics assessment
|
ESR: Susana Proença Nationality: Portugese Host Organization: Universiteit Utrecht, Utrecht, The Netherlands Person in charge: Ass. Prof. Nynke Kramer email: N.I.Kramer@uu.nl; s.proenca@uu.nl |
 |
Susana is currently evaluating current literature models and considerations for in vitro biokinetic assessment. This review will indicate the optimal biokinetic model for the several iPSCs models developed in this project. Simultaneously, biokinetic information (e.g. enzymes involved in the metabolism, possible transporters, specific affinities to cellular organelles) is being collected from literature for an initial set of chemicals chosen to evaluate/characaterized the in vitro models of the project. By integrating the literature information on in vivo buiokinetics, with biokinetic modelling simulations and experimental data on the concentration of the several chemical in medium and cells in time, we will hopefully assess the actual in vitro models sensitiveness. And to compare the intrinsic toxic potencies of a selected group of test chemicals, physiologically based pharmacokinetic (PBPK) models will be developed to estimate human relevant toxic doses from in vitro determined "point of departures", which have been analysed by ESRs at partner institutions. The ESR will make use of existing PBPK modelling platforms such as the population-based PBPK software SimCyp Simulator® with Mech KiM module to simulate internal tissue/cell concentration/time profiles of test chemicals from external exposure regimes for their accumulation potential in the organs of interest, as well as derive steady state plasma concentrations and daily human oral doses from in vitro effect concentrations. Such modelling platforms allow for the comparison of toxic potencies of the iPSC individuals compared to specific population groups.
Potential outcomes:
- Identification of in vitro system components and physicochemical properties affecting the accumulation of test chemicals over time in iPSC systems
- To provide a proof-of-principle strategy to estimate human NOAEL for organ-specific toxic chemicals
- To develop a whole body PBPK simulation of the test compounds from the different donor backgrounds
Project 15: Data management and Bioinformatics
|
ESR: Pranika Singh Nationality: Indian Host Organization: Douglas Connect, Basel, Switzerland Person in charge: Dr. Barry Hardy email: barry.hardy@douglasconnect.com; pranika@douglasconnect.com |
 |
Pranika is involved in creating an in3 data base to host external and project data for easy utilisation by all ESRs. A huge amount of data and knowledge generated by different partners needs to be shared, harmonized, and combined with data coming from publicly-available sources and from literature. It will, therefore make it available for use in modelling and bioinformatics workflows. Bioinformatics workflows can then be used to mine this data to uncover enriched pathways providing information on possible mode of actions, specific biomarkers for the toxicity compared on species level and common toxicity-inducing key events like oxidative stress or DNA damage. This will therefore, facilitate the generation of appropriate reporter lines and the development of AOPs. The project will also be aligned to OpenTox activities, bringing best practice on open source and open standards to the project, hence improving on the current fragmented tool culture, improving sustainability and transparency.
Potential outcomes:
- Creation of an optimised data base with toxicogenomic data from large databases
- Bioinformatic analysis of pre-existing data to support decisions for reporter assay generation
- Enriching AOPs with specific knowledge to make them more understandable and usable
- Application of common statistical rules to project data and analysing it for tissue and donor specific effects
- Genomic, proteomics, metabolomics and related pathway enrichment analysis directed towards AOP development
 |
in3 is funded by the Marie Sklodowska-Curie Action - Innovative Training Network under grant no. 721975. |

